Denaxas Lab
We are a multidisciplinary research lab of data scientists, epidemiologists, software engineers, and clinicians working at the intersection of medicine and computer science. Our mission is to improve human health and healthcare by analyzing electronic health records, administrative data, disease registries, and genomic resources.
Research
Electronic health records

Our lab analyzes electronic health records from millions of individuals using advanced statistical approaches in order to develop a better understanding of the causes and mechanisms of disease. Our research can guide clinicians, policy makers, and researchers on how to formulate differential diagnoses, allocate resources, and target research priorities.
Recent research examples:
Electronic Health Record (EHR) phenotyping
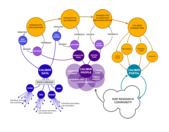
Raw EHR require a substantial amount of preprocessing before they can be transformed into research-ready datasets that can be statistically analyzed to answer clinically meaningful questions. Our lab develops computational algorithms for defining, validating and ascertaining multi-modal disease phenotypes in EHR data. Created phenotypes are stored in the open-access HDR UK Phenotpe Library.
Recent research examples:
Unsupervised machine learning for sub-phenotype discovery
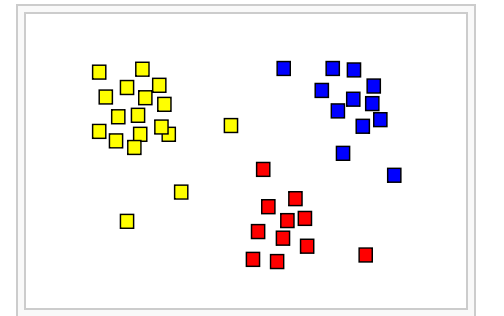
A growing body of evidence from observational and interventional research suggesting that complex diseases, such as type-II diabetes, asthma and chronic obstructive pulmonary disease (COPD), are composed of distinct sub-phenotypes with different risk factor and prognostic profiles. Our lab develops and evaluates unsupervised machine learning algorithms to identify, describe and evaluate disease subtypes that can lead to the development of personalized treatments.
Recent research examples:
Data Linkage
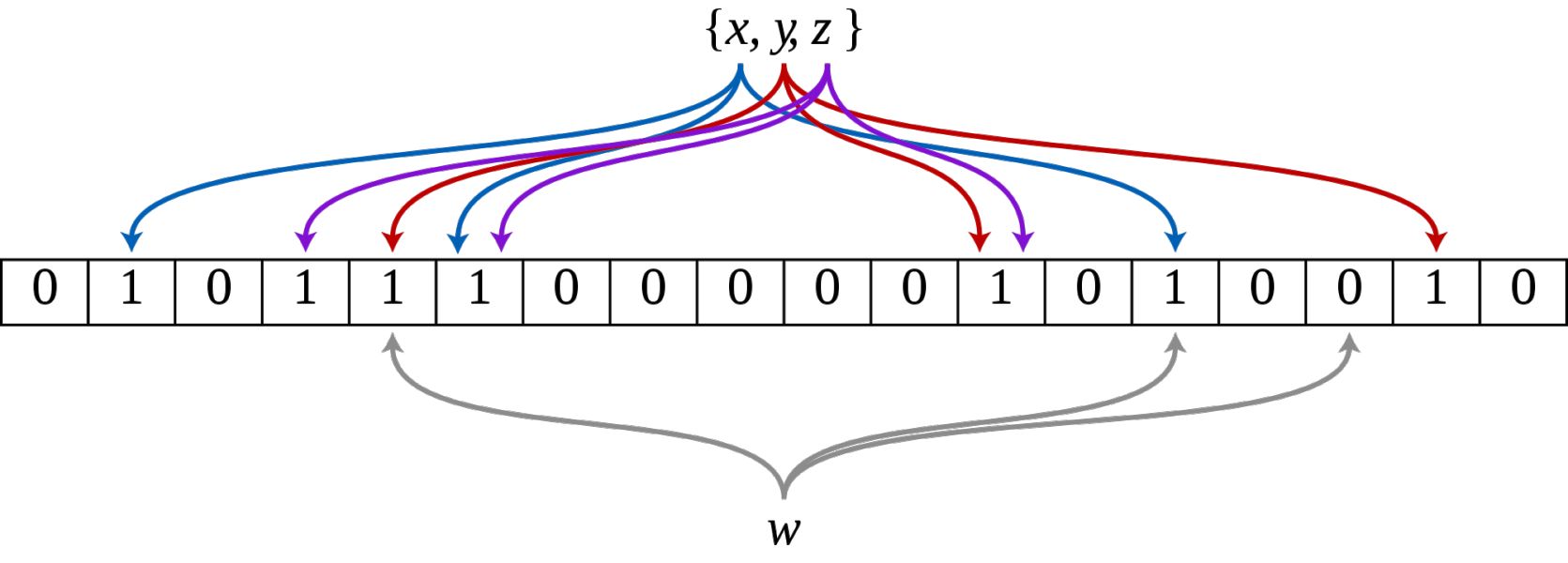
Data linkage is the process of identifying and linking individuals across heterogeneous data sources. Working with the Federal University of Bahia, our lab is contributing to the development of scalable probabilistic data linkage methods for linking administrative over 140 million participants in Brazil and evaluating the quality of the linkage using supervised machine learning.
COVID-19 research initiatives:
Responding to the current public health emergency caused by the SARS-CoV-2 virus and COVID-19 pandemic, our Lab has been actively engaged with several national research initiatives:
- CONVALESCENCE funded by the NIHR and UKRI, this study will analyze data from EHR and longitudinal studies to define and understand long COVID and the impact it has on health and social outcomes.
- COVID-RED is part of the EU Innovative Medicines Initivative and will evaluate the use and performance of a CE-marked device (wearable), which uses sensors to measure breathing rate, pulse rate, skin temperature, and heart rate variability for the purpose of early detection and monitoring of COVID-19 in general and high-risk populations (read trial protocol for more information)
- BHF CVD-COVID-UK aims to understand the relationship between COVID-19 and cardiovascular diseases such as heart attack, heart failure, stroke, and blood clots in the lungs through analyses of de-identified, linked, nationally collated healthcare datasets across the four nations of the UK.
- DECOVID uses detailed and frequently updated health data from hospitals as the COVID-19 pandemic unfolds, to allow clinicians and researchers to generate rapid and robust insights that can lead to more effective clinical treatment strategies, helping patients, healthcare professionals and society.
- BHF COVIDITY-COHORT complements CVD-COVID-UK and will harness the power of over a dozen large UK cohort studies, where participants have already provided a wealth of information about their cardiovascular health, and general health and wellbeing.
Engagement with policy-makers
- Members of the lab performed an analysis and report on community definitions of COVID-19 based on data from symptom trackers for the New and Emerging Respiratory Virus Threats (NERVTAG) and Scientific Advisory Group for Emergencies (SAGE) groups.
- Our research on excess deaths amongst high-risk patients was featured in the UK Parliament Health and Social Care Committee Coronavirus: lessons learnt written evidence report from University College London.
Research output
- Thygesen J. et al. Understanding COVID-19 trajectories from a nationwide linked electronic health record cohort of 56 million people: phenotypes, severity, waves & vaccination. The Lancet Digital Health (accepted), preprint
Gurdasani D. et al. Vaccinating adolescents in England: a risk-benefit analysis. Journal of the Royal Society of Medicine 10.1177/01410768211052589
Media coverage: Daily Mail, The Guardian, BMJ, Royal Society of Medicine, The Times, Financial Times, Daily Mail
- Wood A. et al. Linked electronic health records for research on a nationwide cohort of more than 54 million people in England: data resource. BMJ. 10.1136/bmj.n826
Wilde A. H. et al. The association between mechanical ventilator compatible bed occupancy and mortality risk in intensive care patients with COVID-19: a national retrospective cohort study. BMC Medicine 10.1186/s12916-021-02096-0
Media coverage: Vox, Daily Mail, Independent, Evening Express, BBC News, Yahoo! News.
Eyre M. et al. Impact of baseline cases of cough and fever on UK COVID-19 diagnostic testing rates: estimates from the Bug Watch community cohort study. Wellcome Open Research 10.12688/wellcomeopenres.16304.1
Media coverage: BBC News, The Guardian. Research cited in the Academy of Medical Sciences Preparing for a challenging winter 2020-2021 report.
Banerjee A. et al. Estimating excess 1-year mortality associated with the COVID-19 pandemic according to underlying conditions and age: a population-based cohort study. The Lancet 10.1016/S0140-6736(20)30854-0
Interactive online calculator: OurRisk.Cov
Media coverage: FT, Times, Guardian, BBC, Protagon (GR) and elsewhere.
Lai A. et al. Estimated impact of the COVID-19 pandemic on cancer services and excess 1-year mortality in people with cancer and multimorbidity: near real-time data on cancer care, cancer deaths and a population-based cohort study. BMJ Open 10.1136/bmjopen-2020-043828
Media coverage: The Guardian, Daily Mail, Evening Standard, BMJ.
- Banerjee A. et al. Clinical academic research in the time of Corona: a simulation study in England and a call for action. PLOS ONE 10.1371/journal.pone.0237298
- Dennis J. M. et al. Type 2 Diabetes and COVID-19-Related Mortality in the Critical Care Setting: A National Cohort Study in England, March-July 2020. Diabetes Care 10.2337/dc20-1444
- Katsoulis M. et al. Estimating the effect of reduced attendance at emergency departments for suspected cardiac conditions on cardiac mortality during the COVID-19 pandemic Circulation: Cardiovascular Quality and Outcomes. 10.1161.CIRCOUTCOMES.120.007085
- CVD-COVID-UK Consortium. The 4C Initiative (Clinical Care for Cardiovascular disease in the COVID-19 pandemic): monitoring the indirect impact of the coronavirus pandemic on services for cardiovascular diseases in the UK . 10.1101/2020.07.10.20151118
- Banerjee A. et al. Excess deaths in people with cardiovascular diseases during the COVID-19 pandemic. European Journal of Preventive Cardiology. 10.1093/eurjpc/zwaa155
- Mateen B. et al. A geotemporal survey of hospital bed saturation across England during the first wave of the COVID-19 Pandemic BMJ Open. 10.1136/bmjopen-2020-042945
- Katsoulis M. et al. Obesity during the COVID-19 pandemic: cause of high risk or an effect of lockdown? A population-based electronic health record analysis in 1 958 184 individuals. Public Health. 10.1016/j.puhe.2020.12.003
Funding:
- Awarded funding from the EHDEN COVID-19 Rapid Collaboration Call to transform the UK Biobank to the OHDSI OMOP common data model to facilitate COVID-19 research.
- Funded by the Innovative Medicines Initiative to evaluate wearables for remote diagnosis and monitoring of COVID-19 through the COVID-RED programme.
- Part of large cross-institutional project, the CONVALESCENCE study, funded by the NIHR and UKRI on defining and understanding Long COVID.
Tools
Online calculator for excess deaths due to COVID-19
Using large-scale electronic health records in England, we have developed a simple online tool (OurRisk.Cov) that can calculate and visualize excess deaths over one year from the COVID-19 pandemic based on age, sex, and underlying disease-specific estimates.
Cite HDR UK Phenotype Portal OurRisk.Cov excess mortality risk calculator
Featured Publications
Contact
- s.denaxas[@]ucl.ac.uk
- Institute of Health Informatics, University College London, 222 Euston Road, University College London, London, NW1 2DA
- Institute of Health Informatics